Duration
Description
Ontologies are used in numerous research disciplines and commercial applications to uniformly and semantically annotate real-world objects. Due to a rapid development of application domains the corresponding ontologies are changed frequently to include up-to-date knowledge. These changes dramatically influence dependent data as well as applications/systems, for instance, ontology mappings, that semantically interrelate ontologies. The aim of the work includes the conceptual modelling and the implementation of a framework to systematically study the evolution and its consequences in different domains. Moreover, the robustness of existing match approaches will be analyzed w.r.t. ontology evolution. Based on this, match approaches will be newly designed or optimized.
Basic evolution framework for ontologies and mappings:
Based on a generic framework suitable for analysis of evolution in instance data, ontologies, annotations and ontology mappings (see DILS 2008 paper), we study the evolution 16 currently developed Life Science Ontologies.
Detailed analysis results for 16 currently developed life science ontologies can be accessed here.
GOMMA System:
Generic Ontology Matching and Mapping Management (GOMMA) - See GOMMA project site
GOMMA is our base framework for enhanced applications and analysis listed below.
Ontology Evolution:
COntiDiff / CODEX
To effectively manage the evolution of ontologies it is essential to identify the difference (Diff) between ontology versions. Such a Diff supports the synchronization of changes in collaborative curation, the adaptation of dependent data such as annotations, and ontology version management. We propose a novel approach COnto-Diff to determine an expressive and invertible diff evolution mapping between given versions of an ontology. Our approach first matches the ontology versions and determines an initial evolution mapping consisting of basic change operations (insert/update/delete). To semantically enrich the evolution mapping we adopt a rule-based approach to transform the basic change operations into a smaller set of more complex change operations, such as merge, split, or changes of entire subgraphs. The source code of ContoDiff is available at GitHub. The CODEX (Complex Ontology Diff Explorer) application allows for determining semantic (complex) changes between two versions of an ontology. The web application is based on COntoDiff which applies rules to iteratively compute the most compact (semantically richest) diff between two ontology versions.
OnEX:
OnEX (Ontology Evolution Explorer) supports the exploration of ontology changes to better understand their evolution. \onex is a web-based application that currently provides access up to 500 different versions of 16 well-known life science ontologies including Gene Ontology, NCI Thesaurus and selected OBO ontologies since 2002. The application shows evolution trends of these ontologies and make it possible to study the made changes in detail. More information about OnEX can be found here.
Rex - Discovery of Evolving Ontology Regions:
Ontologies are heavily used in life sciences and evolve continuously to incorporate new or changed insights. Often ontology changes affect only specific parts (regions) of ontologies making it valuable for ontology users and applications to know the heavily changed regions on the one hand and stable regions on the other hand. However, the size and complexity of life science ontologies renders manual approaches to localize changing or stable regions impossible. We therefore propose an approach to automatically discover evolving or stable ontology regions. We evaluate the approach by studying evolving regions in the Gene Ontology and the NCI Thesaurus. More information about the approach and the Region Evolution Explorer (Rex) can be found here: - Region Evolution - DILS 2010. - REX - DILS 2014 - REX - JBS 2015
Efficient versioning of large life science ontologies:
Ontologies have become very popular in life sciences and other domains. They mostly undergo continuous changes and new ontology versions are frequently released. However, current analysis studies do not consider the ontology changes reflected in different versions but typically limit themselves to a specific ontology version which may quickly become obsolete. To allow applications easy access to different ontology versions we propose a central and uniform management of the versions of different biomedical ontologies. The proposed database approach takes concept and structural changes of succeeding ontology versions into account thereby supporting different kinds of change analysis. Furthermore, it is very space-efficient by avoiding redundant storage of ontology components which remain unchanged in different versions. We evaluate the storage requirements and query performance of the proposed approach for the Gene Ontology. More information
Mapping Stability:
Annotation quality considering evolutionary changes:
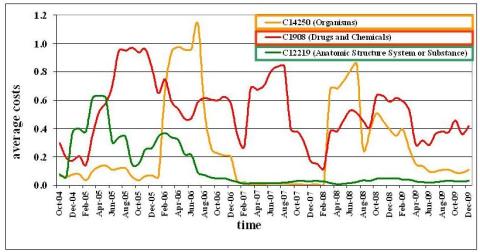
Ontology-based annotations associate objects, such as genes and proteins,with well-defined ontology concepts to semantically and uniformly describe object properties. Such annotation mappings are utilized in different applications and analysis studies whose results strongly depend on the quality of the used annotations. To study the quality of annotations we propose a generic evaluation approach considering the annotation generation methods (provenance) as well as the evolution of ontologies, object sources, and annotations. Thus, it facilitates the identification of reliable annotations, e.g., for use in analysis applications. We evaluate our approach for functional protein annotations in Ensembl and Swiss-Prot using the Gene Ontology. For more information see our DILS 2009 paper
Ontology mapping stability:
Ontology matching has been widely studied. However, the resulting ontology mappings can be rather unstable when the participating ontologies or utilized secondary sources (e.g., instance sources, thesauri) evolve. We propose an evolution-based approach for assessing ontology mappings by annotating their correspondences by information about similarity values for past ontology versions. These annotations allow us to assess the stability of correspondences over time and they can thus be used to determine better and more robust ontology mappings. The approach is generic in that it can be applied independently from the utilized match technique. We define different stability measures and show results of a first evaluation for the life science domain. More information
Publikationen (18)
| Dateien | Cover | Beschreibung | Jahr |
|---|---|---|---|

|
Lin, Y.
; Hoffmann, P.
; Rahm, E.
In Proceedings of the 14th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2021) - Volume 5: HEALTHINF, pages 188-199.
|
2021 / 2 | |

|
Lin, Y.
; Christen, V.
; Groß, A.
; Kirsten, T.
; Cardoso, S.
; Pruski, C.
; Silveira, M.
; Rahm, E.
Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies - Volume 5 HEALTHINF: HEALTHINF, 145-155, 2020, Valletta, Malta. DOI: 10.5220/0008979901450155
|
2020 / 2 | |

|
Christen, V.
; Lin, Y.
; Groß, A.
; Cardoso, S.
; Pruski, C.
; Silveira, M.
; Rahm, E.
Data Integration in the Life Science (DILS) 2018
|
2018 / 11 | |

|
Lin, Y.
; Christen, V.
; Groß, A.
; Cardoso, S.
; Pruski, C.
; Silveira, M.
; Rahm, E.
Proc. Data Integration in the Life Science (DILS) 2017
|
2017 / 11 | |

|
Christen, V.
; Groß, A.
; Fisher, J.
; Wang, Q.
; Christen, P.
; Rahm, E.
Proc. 19th Int. Conf. on Extending Database Technology (EDBT), Venice, 2017
|
2017 / 3 | |

|
Cardoso, S.
; Reynaud-Delaître, C.
; Silveira, M.
; Lin, Y.
; Groß, A.
; Rahm, E.
; Pruski, C.
Proc. 10th Int. Joint Conf.on Biomedical Engineering Systems and Technologies (BIOSTEC 2017), HEALTHINF, Porto, Feb. 2017
|
2017 / 2 | |

|
Cardoso, S.
; Pruski, C.
; Silveira, M.
; Lin, Y.
; Groß, A.
; Rahm, E.
; Reynaud-Delaître, C.
Knowledge Engineering and Knowledge Management. Proc. EKAW, Springer LNCS 10024 , pp. 68-82, 2016
|
2016 / 11 | |

|
Christen, V.
; Groß, A.
; Rahm, E.
Proc. 15th International Semantic Web Conference (ISWC), Springer LNCS 9981, pp. 135-150 , 2016
|
2016 / 10 | |

|
Groß, A.
; Hartung, M.
; Kirsten, T.
; Rahm, E.
2nd International Conference on Biomedical Ontology (ICBO 2011)
|
2011 / 7 | |

|
Groß, A.
; Hartung, M.
; Kirsten, T.
; Rahm, E.
7th International Conference on Data Integration in the Life Sciences (DILS 2010)
|
2010 / 8 |